Gene study identifies series of DNA variants linked to dyslexia
Scientists from the Universities of St Andrews and Edinburgh have, for the first time, pinpointed a large number of genes that are reliably associated with dyslexia.
Around a third of the 42 genetic variants identified have been previously linked to general cognitive ability and educational attainment.
The researchers say their findings, published in Nature Genetics journal today (Thursday 20 October) aid our understanding of the biology behind why some children struggle to read or spell.
Dyslexia is known to run in families – partly because of genetic factors – but, until now, little was known about the specific genes that relate to the risk of it developing.
The study, led by the University of Edinburgh alongside researchers from the University of St Andrews School of Medicine, is the largest genetic study of dyslexia to date. Previous studies linking dyslexia to specific genes have been done on small numbers of families and the evidence was unclear, the research team says.
This latest study involved more than 50,000 adults who have been diagnosed with dyslexia and more than one million adults who have not.
Researchers tested the association between millions of genetic variants with dyslexia status and found 42 significant variants.
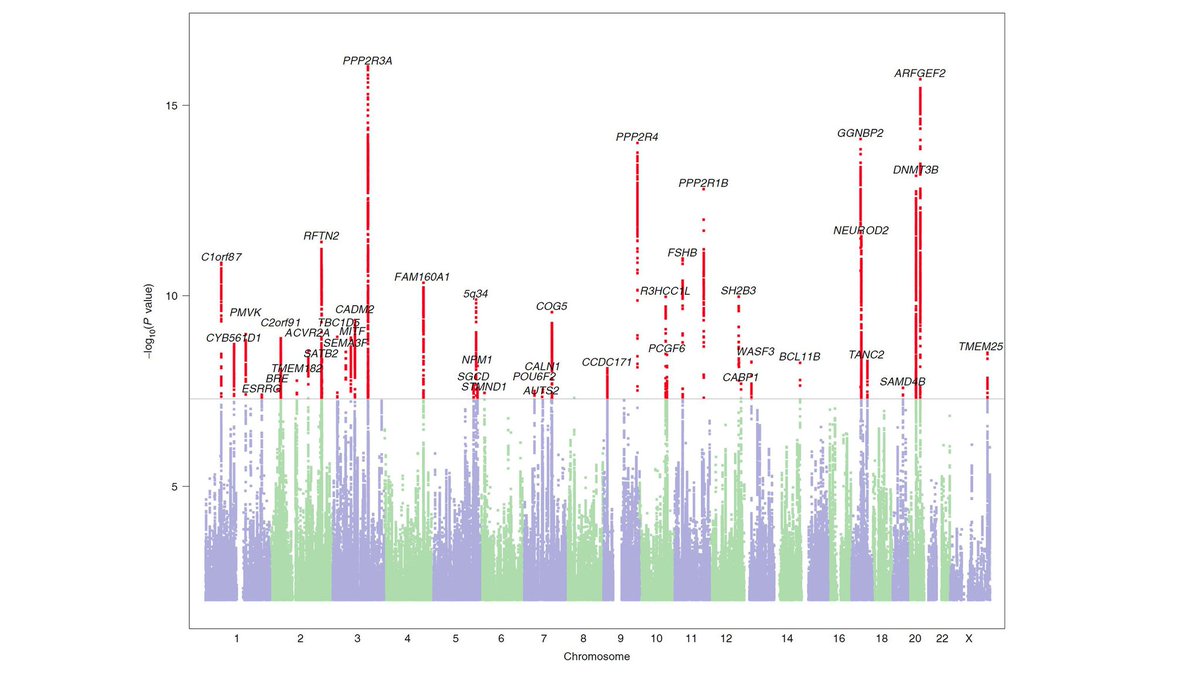
Some of these are associated with other neurodevelopment conditions, such as language delay, and with thinking skills and academic achievement. Many, however, are novel and could represent genes that more specifically associate with processes essential for learning to read.
Many of the genes associated with dyslexia are also associated with attention deficit hyperactivity disorder. A much smaller overlap of the genes associated with dyslexia was found for psychiatric, lifestyle and health conditions.
Several of the associated genetic variants were also significant in a Chinese speaking sample suggesting that there are general cognitive processes in learning to read that are not dependent on the type of language.
Researchers say they were able to predict how well children and adults from four other research studies can read and spell using the genetic information from the study, but not with the accuracy needed for diagnostic use.
Other key researchers in the study were from the Max Planck Institute for Psycholinguistics in the Netherlands, QIMR Berghofer Medical Research Institute in Australia and US company 23andMe, Inc.
Lead researcher Michelle Luciano, of the University of Edinburgh’s School of Philosophy, Psychology and Language Sciences, says the study sheds light on many unanswered questions around dyslexia.
“Our findings show that common genetic differences have very similar effects in boys and girls, and that there is a genetic link between dyslexia and ambidexterity,” says Dr Luciano. “Previous work suggested some brain structures may be altered in people with dyslexia, but we did not find evidence that genes explain this.”
“Our results also suggest that dyslexia is very closely genetically related to performance on reading and spelling tests reinforcing the importance of standardised testing in identifying dyslexia.”
“This is a breakthrough in the field,” says Dr Silvia Paracchini from the University of St Andrews and co-author on the study, “and I am delighted to work with Michelle to make this work relevant for people with dyslexia. We have set-up the Specific Learning Difficulties Network (SLDN) bringing together researchers and the key stakeholders to redefine the research agenda of this field.”
Category Research