Discovery of bacterial enzyme structure could lead to new antibiotics
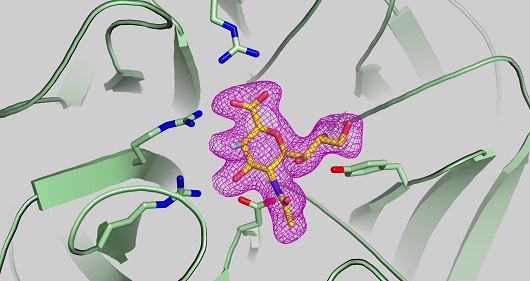
Scientists from the University of St Andrews and Diamond Light Source have revealed the structure of a key enzyme in Streptococcus pneumonia, according to research published in the Journal of Biological Chemistry; paving the way to new treatments for a range of respiratory infections including pneumonia, meningitis and septicaemia.
Respiratory infections are one of the biggest killers in the western world, and pneumococcal infections can range from mild to potentially deadly. Children and older people are most vulnerable, as well as those with compromised immune systems.
Scientists from Scotland’s first University used the UK’s synchrotron science facility, Diamond Light Source, to uncover the structure of NanC, a type of enzyme known as a ‘neuraminidase’. Using crystallographic techniques on the synchrotron’s MX beamlines they successfully unlocked the secrets of the enzymes that help the bacteria to grow and spread – an important target for future antibiotics.
Streptococcus pneumoniae bacteria contain up to three neuraminidase enzymes: Nan A, B and C. The scientists had already uncovered the structure of the other two neuraminidases – A and B – and the discovery of the NanC structure is the final step in identifying how these enzymes work to sustain S. pneumoniae and how they could be targeted and disabled, thus destroying the bacteria.
 Martin Walsh, deputy life sciences director at Diamond and lead author on the study, said: “With the structure of NanC, we now have a vital new piece of the puzzle which informs wider research into drug design and equips us to better tackle the pneumococcus in all its forms. The next step will be for scientists to use this structural information to aid in the design of inhibitors of the pneumococcus neuraminidases which could lead to new treatments for pneumococcal disease.”
Martin Walsh, deputy life sciences director at Diamond and lead author on the study, said: “With the structure of NanC, we now have a vital new piece of the puzzle which informs wider research into drug design and equips us to better tackle the pneumococcus in all its forms. The next step will be for scientists to use this structural information to aid in the design of inhibitors of the pneumococcus neuraminidases which could lead to new treatments for pneumococcal disease.”
There are approximately 90 distinct pneumococcal serotypes and, although not all cause disease, this makes treatment challenging. An effective conjugate vaccine has been available since 2001, which targets seven serotypes and has significantly reduced the invasive pneumococcal disease burden in the first world, especially for the vulnerable. This has recently been augmented by the introduction of a 13 serotype conjugate vaccine.
However there has been a rise in antibiotic resistance in serotypes not covered by this conjugate vaccine and previously non-infectious serotypes are increasing in virulence. Earlier this month the World Health Organization warned the world is heading for a “post-antibiotic era” unless new drugs are developed. The chief medical officer for England Professor Dame Sally Davies evoked parallels with the “apocalypse” in calls for a renewed focus on developing new antibiotics. Finding new approaches to combat pneumococcal disease is a vital contribution to this effort.
Because the pneumococcus rely on neuraminidases for survival, drugs that shut off these enzymes could aid new ways to treat infections. This approach is already used for influenza virus: both Tamiflu and Relenza target influenza neuraminidase, which prevents the virus infection spreading. No equivalent drugs exist, as yet, for bacterial infections.
The group’s findings show that NanC does not function exactly like Nan A and B, so future antibiotics will have to work broadly enough to attack all three neuraminidases.
Garry Taylor, Professor of Molecular Biophysics and Deputy Principal of the University of St Andrews, said: “With a fuller understanding of the structure and function of S. pnemoniae’s three neuraminidase enzymes, we’re now better equipped to design next-generation antibiotics for some of the world’s most prevalent and deadly respiratory infections.”

Notes to news editors
This research was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) and the Medical Research Council (MRC).
The full paper is published in the Journal of Biological Chemistry. DOI: 10.1074/jbc.M115.673632
Image credits:
– A sugar-mimicking inhibitor molecule bound to the NanC protein; credit: Petra Lukacik, Diamond Light Source
– The high tech machinery on MX beamline I04; courtesy of Diamond Light Source
– Diamond Light Source aerial view; courtesy of Diamond Light Source
Issued by the University of St Andrews Press Office: contactable on [email protected] or 01334 46 2530.
Category Research